open metadata Markup Language - Home

odML is an initiative to define and establish an open, flexible and easy-to-use format to transport metadata. We started it in the first place to connect our two software projects RELACS (by Jan Benda) for acquiring electrophysiological data and the LabLog (by Jan Grewe) for project documentation and (meta)data management.
See recent paper: Grewe J, Wachtler T and Benda J (2011) A bottom-up approach to data annotation in neurophysiology. Front. Neuroinform. 5:16
Metadata is data about data, i.e. it describes the conditions under which the actual raw-data were acquired.
What does that mean? A simple example in the electrophysiological context would be the sample rate with which the acquired data has been recorded. This may sound ridiculous for everyone doing such an experiment should know how the data was acquired. But what if the data has to be shared in a collaboration? At least at this point this little piece of information is essential to make sense out of the shared data, it must not get lost over time. Of course, a complete description about the data includes much more e.g. the stimulus parameters, ambient conditions, etc...
Most of this information is known by the programs used for data acquisition and stimulation. So far, almost any one vendor of acquisition software uses its own format with which such metadata is saved along with the raw-data. Thus there are many different formats around which makes it hard to retrieve the information from files a different tool has written. We hope odML can help to ease this. Here, we describe our approach to this issue.
Talking about data
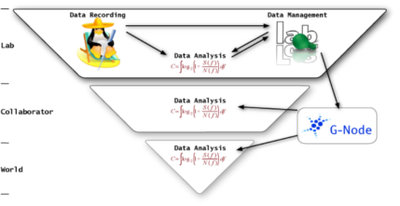
The figure illustrates the Food-Chain of science. At its basis (on top) is the lab in which data is recorded, analysed and managed. All the arrows indicate the flow of data and metadata. As said above we need metadata in order to analyse and interpret experimental data. If it comes to sharing of data with collaborators or also with the world (e.g. via public dataservers as CRCNS or G-Node et al.) We need not only to pass the actual data but also lots of information about the applied stimuli, the settings of applied filters etc., i.e. metadata.
Storing metadata increases the data lifecycle
Unfortunately the knowlegde about once aquired data has the tendency to vanish with time. Only well annotated data can be easily reused. Annotation should start as early as possible in the data lifecylce ideally right at the time of recording and should be as far automated as possible.
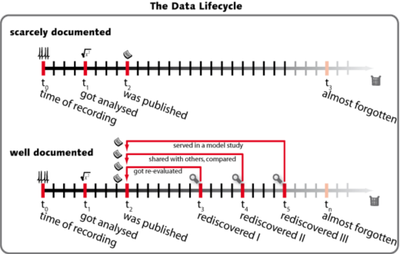
Anotating data and storing metadata can increase the lifetime of acquired data! Your data deserves it.
For annotation and sharing of data it is necessary to have a format that fulfills certain requirements:
- Easy to use and ideally human readable.
- Can be implemented into any recording, analysis or management tool.
- Open and freely available.
- Inherently extensible and flexible for science continually changes.
- More or less unresticted i.e. it must/should not restrict the user or strictly require entries.
Related initiatives
The issue of sharing data or keeping data publicly available in databases has raised the problem of defining metadata neccesary to make sense out of the data. The first initiative in the neuroscience community heading towards this is the so called MINI by the CARMEN project. We included their "Mimimum information about a Neuroscience Investiagtion" (see their paper) to be part of our terminologies.